Research sheds new light on unusual DNA structures likely contributing to the risk of osteoporosis
|Osteoporosis is the most common chronic metabolic bone disease, characterized by increased bone fragility. According to the International Osteoporosis Foundation, it causes more than 8.9 million fractures annually worldwide. Pathogenic conditions that lead to osteoporosis disrupt the tightly regulated balance between bone resorption and formation. One of the key regulators of bone homeostasis is RANKL protein, whose expression is considerably upregulated in osteoporosis. Molecular mechanisms by which expression of a RANKL gene could be regulated are not fully explained yet. A study published by CERIC users in 2018, aimed to uncover new therapeutic strategies, which could lead to more efficient osteoporosis treatments.
One of the potential ways of gene regulation on the transcriptional level is by the formation of unusual secondary DNA structures that are rich in guanine (i.e. one of the nitrogen-containing bases composing the DNA, together with A-adenine, T-thymine and C-cytosine), such as G-quadruplexes (G4), in the regulatory regions of a particular gene. The most characteristic structural element of G4 is G-quartet, a planar arrangement of four guanines. G-quartets stack on top of each other to form the G4 core, while residues of other bases, which connect guanine planes, form so-called “loops” of different sequences, length, and orientations. Formation of G4 in gene regulatory regions, where they can act as activators or repressors of gene transcription, was so far connected with diseases such as cancer and neurodegenerative disorders.
For the first time, a team of researchers including Martina Lenarčič Živković, Jan Rozman and prof. Janez Plavec, from Slovenian NMR Centre in Ljubljana, showed that G4 structures can form in the RANKL gene, considered responsible for the development of osteoporosis. In particular, they observed that the G-rich DNA sequence d(GGGTAGGGAGCGGGAGAGGG) from the RANKL gene regulatory region (RAN4), folds into a G4 with only two G-quartets (fig. 1), although a three-quartet G4 (i.e., three G-quartet stacks, as in fig. 2) was expected to form based on the RAN4 sequence that posseses four GGG tracts. In contrast to protein structures, which can be more accurately predicted based on the sequence, predictions of G4 structures are not so reliable, especially due to the diverse roles of loop residues in the process of folding (they can, for example, either stabilize or prevent the formation of particular structures). How a single loop residue can influence the final structure that a particular G-rich sequence will occupy, was clearly shown on the case of RAN4. The analysis of the RAN4 high-resolution structure determined with NMR spectroscopy at the Slovenian partner facility of CERIC, the NMR Centre at the National Institute of Chemistry in Ljubljana, uncovered the decisive role of a single loop adenine A5 which, by forming a base triad outside the quartet, led to a distinctive two-quartet G4 structure.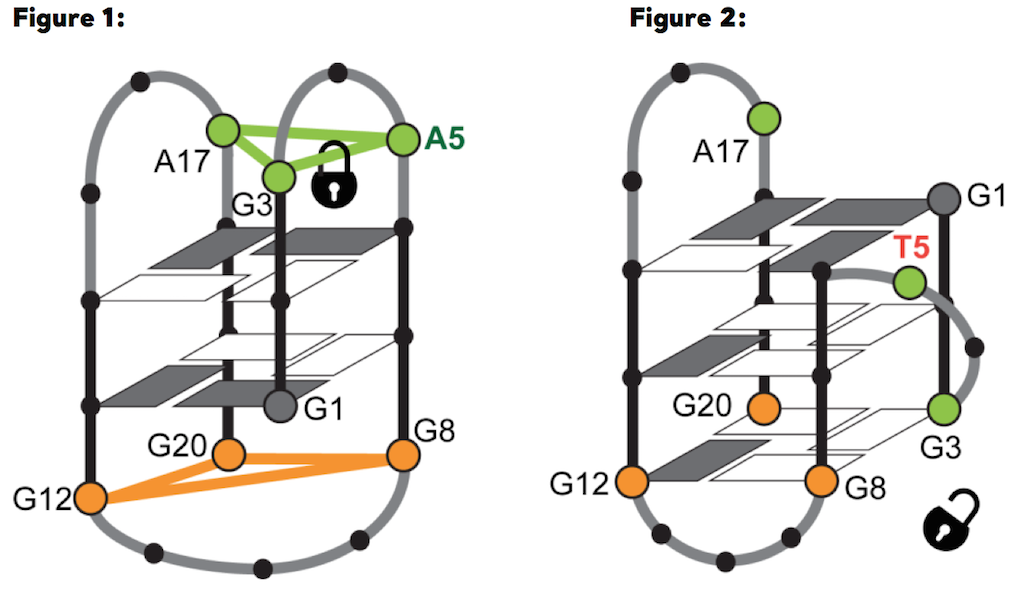 Structural switch between two-quartet (left [a], RAN4) and three-quartet (right [b], RAN4A5T) G-quadruplex depends on interactions of adenine A5 from A•G•A base-triad, where it forms hydrogen bonds with G3
Structural switch between two-quartet (left [a], RAN4) and three-quartet (right [b], RAN4A5T) G-quadruplex depends on interactions of adenine A5 from A•G•A base-triad, where it forms hydrogen bonds with G3
These results indicate that specific loop interactions involving an adenine residue can critically influence the structure of a G4 and provide insights into the complexity of intricate interactions that can guide the folding process of G-rich sequences. The study shows that RAN4 sequence is able to form G4 structure(s) in vitro. However, the next step of the research will focus on the evaluation of formation and potential influence of the observed structure in the cell. The study is the first to suggest that expression of the RANKL gene may be regulated by folding of its G-rich region into unusual DNA structures, such as G4, and as such, represents a novel idea towards the development of potential G4-mediated osteoporosis therapeutic strategies.



