NMR analyses shed light on non-canonical 3D DNA structures
|It is well known that, in addition to the famous double helix structure described by Watson and Crick in 1953, certain genomic segments can adopt a variety of peculiar 3D structures. Formation of such non-canonical architectures appears more frequent in regions comprising tandem repeats and is promoted by particular internal or environmental stimuli.
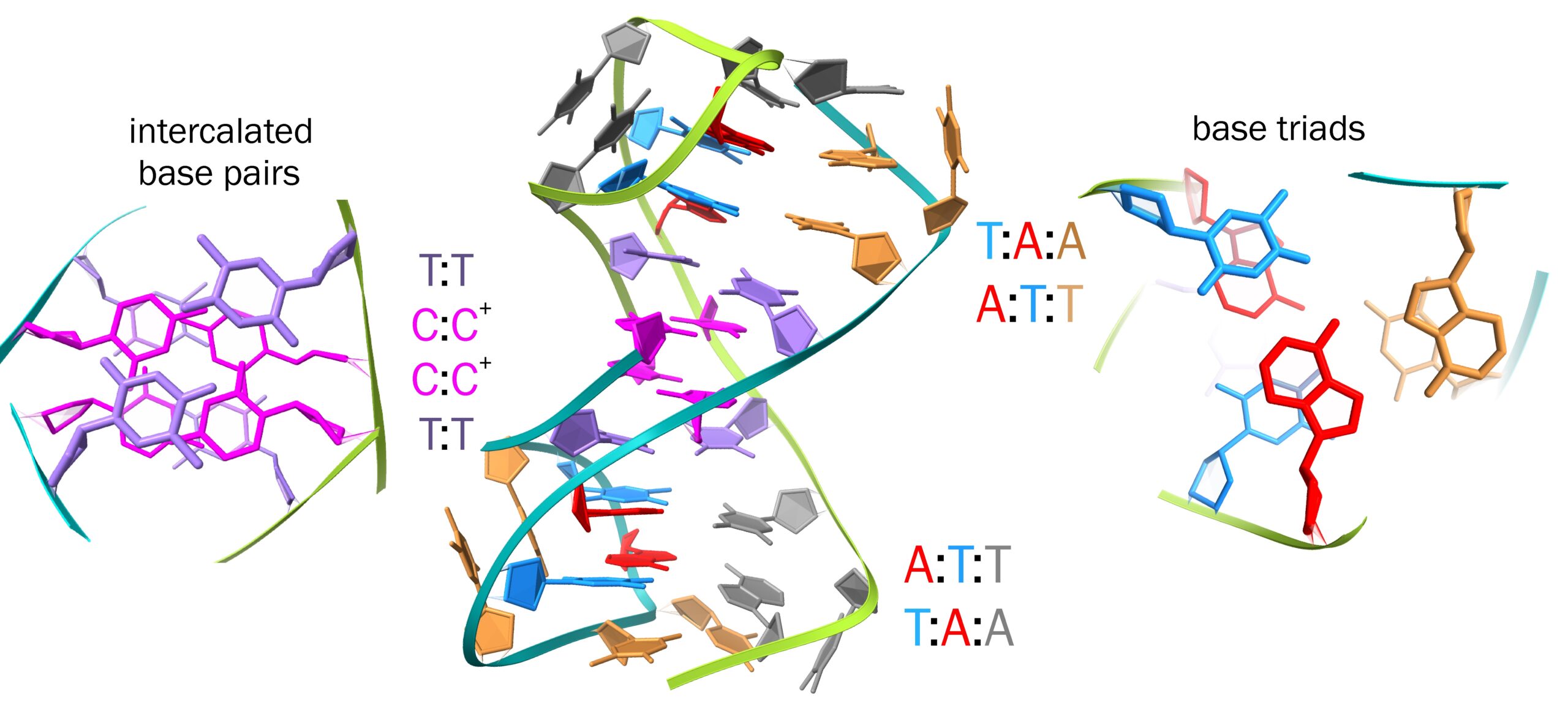
Dr. Marko Trajkovski (Slovenian NMR Centre, National Institute of Chemistry), Prof. Annalisa Pastore (King’s College London), and Prof. Janez Plavec (Slovenian NMR Centre) analysed and characterised DNA oligonucleotides comprising repeats of five nucleotides, ATTTC, that are present in hundreds of genes which play crucial roles in several physiological cellular processes, such as transcriptional regulation, intracellular signalling, protein and membrane trafficking and cell adhesion. At the same time, the insertion and expansion of this pentanucleotide is observed in non-coding genomic regions associated with a number of neurological disorders (including some subtypes of ataxia and epilepsy).
Exploiting advanced Nuclear Magnetic Resonance instruments available at the Slovenian CERIC Partner Facility in Ljubljana, such as DAVID and ASKA spectrometers, scientists showed that, in a solution containing divalent cations, such as Mg2+ and Ca2+, DNA comprising ATTTC repeats folds into a novel architecture: in this 3D structure, the central part corresponds to a block of four intercalated C:C+ and T:T base pairs, and is extended on each side by segments stabilised by pairs of base triads.
These results could enhance the understanding of the physiological and pathological roles of non-canonical 3D structures, helping to describe how these processes are regulated in living cells.
ORIGINAL ARTICLE:



